Analyzing Your Patients with NGP
Next-Generation Phenotyping
Face2Gene™ is proud to present different algorithms and visualizations, to provide you with state-of-the-art NGP analysis for your patients
* Currently, the magnitude of similarities is not calibrated and does not allow comparison between the different algorithms*
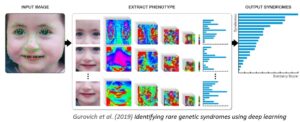
DeepGestalt™
The input photo is first pre-processed to achieve facial detection, landmark detection and alignment.
After preprocessing, the input image is cropped into facial regions. Each region is fed into a Deep Convolutional Neural Network (DCNN) to obtain a softmax vector indicating its correspondence to each syndrome in the model. The output vectors of all regional DCNNs are then aggregated and sorted to obtain a final ranked list of genetic syndromes — the 30 syndrome matches displayed in Face2Gene™ CLINIC’s RARE tab. More details in Nature Medicine. Current availability: web, iOS & Android app.
FeatureMatcher
Following the Human Phenotype Ontology terms, this algorithm allows a phenotype-based syndrome prioritization. In Face2Gene ‘s RARE tab, these results are combined with DeepGestalt’s results to provide a ranked list of syndromes. More details on how this was used in PEDIA algorithm, in Genetics in Medicine. FeatureMatcher is also available in LIBRARY and Face2Gene™ LABS. Current availability of CLINIC: web, iOS & Android app.
GestaltMatcher™
Based on the DeepGestalt framework a “Clinical Face Phenotype Space” is created, such that the distance between photos defines syndromic similarity. This is listed in Face2Gene CLINIC’s ULTRA-RARE tab, allowing patient photos to be matched to a molecular diagnosis even when the disorder was not part of the training set. The output is displayed in two parallel lists, the first one ranking the matched patients, and the second one, ranking the syndromes of these matched patients.
Similarities among patients with previously unknown disease genes can also be detected, and these are displayed in the UNDIAGNOSED tab in Face2Gene CLINIC. More details in Nature Genetics. Current availability: web app only.
Facial D-score
Based on the algorithms described above we built descriptors to differentiate between 2 classes of frontal facial photos: images of patients diagnosed with a rare genetic disease and presenting a facial dysmorphia, and an equivalently sampled second class of images of unaffected individuals. The tool currently supports pediatric-aged patients and is the algorithm that powers the “Pediatrician View” in Face2Gene. More information in this preliminary study as well as this JMIR publication. Current availability: web and mobile apps.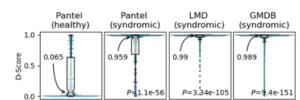
GeneSearch
Combining DeepGestalt, FeatureMatcher and extensive gene mutation databases, this algorithm enables correlation of NGS results with all the phenotypic information captured by the above mentioned algorithms in Face2Gene CLINIC, thus facilitating the more accurate and efficient interpretation of genomic variant profiles. More details on how this was used in the PEDIA algorithm, in Genetics in Medicine. Current availability: Face2Gene LAB web only.
Text2Phenotype Beta
Text-mined phenotype annotation allows extracting the features from clinical notes directly into Human Phenotype Ontology terms. Available also as dictated text, these terms can be then analyzed by the FeatureMatcher algorithm and combined with DeepGestalt to render a ranked list of syndrome matches listed in Face2Gene CLINIC’s RARE tab. Current availability: Face2GeneCLINIC web and mobile apps under “Analyze Clinical Note”, as well as Face2Gene LAB.
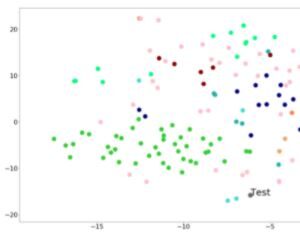
tSNE visualization
Shows a 2D projection of the case image into a “Clinical Face Phenotype Space” as compared to relevant syndromes. There are currently 2 TSNE visualizations available in Face2Gene CLINIC: the first one providing a full analysis for your patient under “Graphical View”, and a second one, for the matched undiagnosed patient. The first tSNE displays the projection of your patient’s photo as compared to the top-10 syndromes analyzed by the DeepGestalt and GestaltMatcher algorithms*. The second tSNE, in the UNDIAGNOSED tab, projects the matched photo as compared to the top-10 syndromes resulting from the GestaltMatcher algorithm analysis. Current availability: Face2Gene CLINIC web & mobile apps.

Masks/ composite images
The facial descriptors can also be graphically displayed as a two-dimensional model of the face specific to the particular syndrome of interest. These composites are created taking an average of the images that participated in the training of a particular syndrome.
Current availability: Face2Gene CLINIC web, iOS & Android apps

Heatmaps
A graphical heatmap can be applied to visualize the degree of similarity between the two descriptors being compared, namely, the descriptor of the patient’s photo being analyzed and the composite image of the syndrome being studied.
Current availability: Face2Gene CLINIC web, iOS & Android apps
* Currently, the magnitude of similarities is not calibrated. Please do not compare between the magnitudes of results of the different algorithms.